Plots temporal variation for different variables, typically pollutant concentrations, across user-defined time scales. Multiple panels can be shown, such as hour of the day, day of the week, week of the year, month of the year, annual mean, or any other time-based grouping the user specifies. By default, this function plots the diurnal, day of the week and monthly variation for different variables, typically pollutant concentrations. Four separate plots are produced.
Usage
timeVariation(
mydata,
pollutant = "nox",
panels = c("hour.weekday", "hour", "month", "weekday"),
local.tz = NULL,
normalise = FALSE,
xlab = NULL,
name.pol = pollutant,
type = "default",
group = NULL,
difference = FALSE,
statistic = "mean",
conf.int = 0.95,
B = 100,
ci = TRUE,
cols = "hue",
ref.y = NULL,
key = NULL,
key.columns = NULL,
start.day = 1,
panel.gap = 0.2,
auto.text = TRUE,
alpha = 0.4,
plot = TRUE,
...
)Arguments
- mydata
A data frame of time series. Must include a
datefield and at least one variable to plot.- pollutant
Name of variable to plot. Two or more pollutants can be plotted, in which case a form like
pollutant = c("nox", "co")should be used.- panels
A vector of character values which can be passed to
cutData(); used to define each panel in the plot. The first panel will take up the entire first row, and any remaining panels will make up the bottom row. If a single panel is given, it will take up the entire plotting area. Combining twotypestrings delimited with a full stop (e.g.,"hour.weekday") will use the first as the x-axis variable the second as a facet.- local.tz
Should the results be calculated in local time that includes a treatment of daylight savings time (DST)? The default is not to consider DST issues, provided the data were imported without a DST offset. Emissions activity tends to occur at local time e.g. rush hour is at 8 am every day. When the clocks go forward in spring, the emissions are effectively released into the atmosphere typically 1 hour earlier during the summertime i.e. when DST applies. When plotting diurnal profiles, this has the effect of “smearing-out” the concentrations. Sometimes, a useful approach is to express time as local time. This correction tends to produce better-defined diurnal profiles of concentration (or other variables) and allows a better comparison to be made with emissions/activity data. If set to
FALSEthen GMT is used. Examples of usage includelocal.tz = "Europe/London",local.tz = "America/New_York". SeecutDataandimportfor more details.- normalise
Should variables be normalised? The default is
FALSE. IfTRUEthen the variable(s) are divided by their mean values. This helps to compare the shape of the diurnal trends for variables on very different scales.- xlab
x-axis label; one for each
panel. Defaults to the x-axis variable defined inpanels. Must be the same length aspanels.- name.pol
This option can be used to give alternative names for the variables plotted. Instead of taking the column headings as names, the user can supply replacements. For example, if a column had the name "nox" and the user wanted a different description, then setting
name.pol = "nox before change"can be used. If more than one pollutant is plotted then usece.g.name.pol = c("nox here", "o3 there").- type
typedetermines how the data are split i.e. conditioned, and then plotted. The default is will produce a single plot using the entire data. Type can be one of the built-in types as detailed incutData(), e.g.,"season","year","weekday"and so on. For example,type = "season"will produce four plots — one for each season.It is also possible to choose
typeas another variable in the data frame. If that variable is numeric, then the data will be split into four quantiles (if possible) and labelled accordingly. If type is an existing character or factor variable, then those categories/levels will be used directly. This offers great flexibility for understanding the variation of different variables and how they depend on one another.Only one
typeis allowed intimeVariation(), and it is applied to eachpanel. For additional splits, use the"x.type"syntax in thepanelsargument (e.g,panels = c("hour.weekday")).- group
This sets the grouping variable to be used. For example, if a data frame had a column
sitesettinggroup = "site"will plot all sites together in each panel.- difference
If two pollutants are chosen then setting
difference = TRUEwill also plot the difference in means between the two variables aspollutant[2] - pollutant[1]. Bootstrap 95\ difference in means are also calculated. A horizontal dashed line is shown at y = 0. The difference can also be calculated if there is a column that identifies two groups, e.g., having usedsplitByDate(). In this case it is possible to calltimeVariation()with the optiongroup = "split.by"anddifference = TRUE.- statistic
Can be
"mean"(default) or"median". If the statistic is"mean"then the mean line and the 95% confidence interval in the mean are plotted by default. If the statistic is"median"then the median line is plotted together with the 5/95 and 25/75th quantiles are plotted. Users can control the confidence intervals withconf.int.- conf.int
The confidence intervals to be plotted. If
statistic = "mean"then the confidence intervals in the mean are plotted. Ifstatistic = "median"then theconf.intand1 - conf.intquantiles are plotted.conf.intcan be of length 2, which is most useful for showing quantiles. For exampleconf.int = c(0.75, 0.99)will yield a plot showing the median, 25/75 and 5/95th quantiles.- B
Number of bootstrap replicates to use. Can be useful to reduce this value when there are a large number of observations available to increase the speed of the calculations without affecting the 95% confidence interval calculations by much.
- ci
Should confidence intervals be shown? The default is
TRUE. Setting this toFALSEcan be useful if multiple pollutants are chosen where over-lapping confidence intervals can over complicate plots.- cols
Colours to be used for plotting; see
openColours()for details.- ref.y
A list with details of the horizontal lines to be added representing reference line(s). For example,
ref.y = list(h = 50, lty = 5)will add a dashed horizontal line at 50. Several lines can be plotted e.g.ref.y = list(h = c(50, 100), lty = c(1, 5), col = c("green", "blue")). Seepanel.ablinein thelatticepackage for more details on adding/controlling lines.- key
By default
timeVariation()produces four plots on one page. While it is useful to see these plots together, it is sometimes necessary just to use one for a report. IfkeyisTRUE, a key is added to all plots allowing the extraction of a single plot with key. See below for an example. IfkeyisFALSE, no key is shown for any plot.- key.columns
Number of columns to be used in the key. With many pollutants a single column can make to key too wide. The user can thus choose to use several columns by setting
columnsto be less than the number of pollutants.- start.day
What day of the week should the plots start on? The user can change the start day by supplying an integer between
0and6.Sunday = 0,Monday = 1, and so on. For example to start the weekday plots on a Saturday, choosestart.day = 6.- panel.gap
The gap between panels in any split panel (e.g., the default
"hour.weekday"panel).- auto.text
Either
TRUE(default) orFALSE. IfTRUEtitles and axis labels will automatically try and format pollutant names and units properly, e.g., by subscripting the '2' in NO2.- alpha
The alpha transparency used for plotting confidence intervals.
0is fully transparent and 1 is opaque. The default is0.4.- plot
Should a plot be produced?
FALSEcan be useful when analysing data to extract plot components and plotting them in other ways.- ...
Other graphical parameters passed onto
lattice::xyplot()andcutData(). For example, in the case ofcutData()the optionhemisphere = "southern". Note thatcutData()is used intype,groupandpanels, and...will be passed to all three.
Value
an openair object. The components of
timeVariation() are named after panels. Associated data.frames can be
extracted directly using the subset option, e.g. as in plot(object, subset = "day.hour"), summary(output, subset = "hour"), etc., for
output <- timeVariation(mydata, "nox")
Details
The variation of pollutant concentrations by time can reveal many interesting features that relate to source types and meteorology. For traffic sources, there are often important differences in the way vehicles vary by type - e.g., fewer heavy vehicles at weekends.
The timeVariation() function makes it easy to see how concentrations (and
many other variable types) vary across different temporal resolutions. Users
have full control over which based panels are shown, allowing for more
tailored and insightful analysis.
The plots also show the 95% confidence intervals in the mean. The 95% confidence intervals are calculated through bootstrap simulations, which will provide more robust estimates of the confidence intervals (particularly when there are relatively few data).
The function can handle multiple pollutants and uses the flexible type
option to provide separate panels for each 'type' — see cutData() for more
details. timeVariation() also accepts a group option, useful for stacked
data. This works similarly to having multiple pollutants in separate columns.
Users can supply their own ylim, e.g. ylim = c(0, 200), which will be
used for all plots. Alternatively, ylim can be a list equal to the length
of panels to control y-limits for each individual panel, e.g. ylim = list(c(-100,500), c(200, 300), c(-400,400), c(50,70)).
The difference option calculates the difference in means between two
pollutants, along with bootstrap estimates of the 95\
in the difference. This works in two ways: either two pollutants are supplied
in separate columns (e.g. pollutant = c("no2", "o3")), or there are two
unique values of group. The difference is calculated as the second
pollutant minus the first and is labelled accordingly. This feature is
particularly useful for model evaluation and identifying where models diverge
from observations across time scales.
Note also that the timeVariation() function works well on a subset of data
and in conjunction with other plots. For example, a polarPlot() may
highlight an interesting feature for a particular wind speed/direction range.
By filtering for those conditions timeVariation() can help determine
whether the temporal variation of that feature differs from other features
— and help with source identification.
The function also supports non-pollutant variables, such as meteorological or traffic flow data.
Depending on the choice of statistic, a subheading is added. Users can
control the text in the subheading through the use of sub e.g. sub = ""
will remove any subheading.
See also
Other time series and trend functions:
TheilSen(),
calendarPlot(),
runRegression(),
smoothTrend(),
timePlot(),
timeProp(),
trendLevel()
Examples
# basic use
timeVariation(mydata, pollutant = "nox")
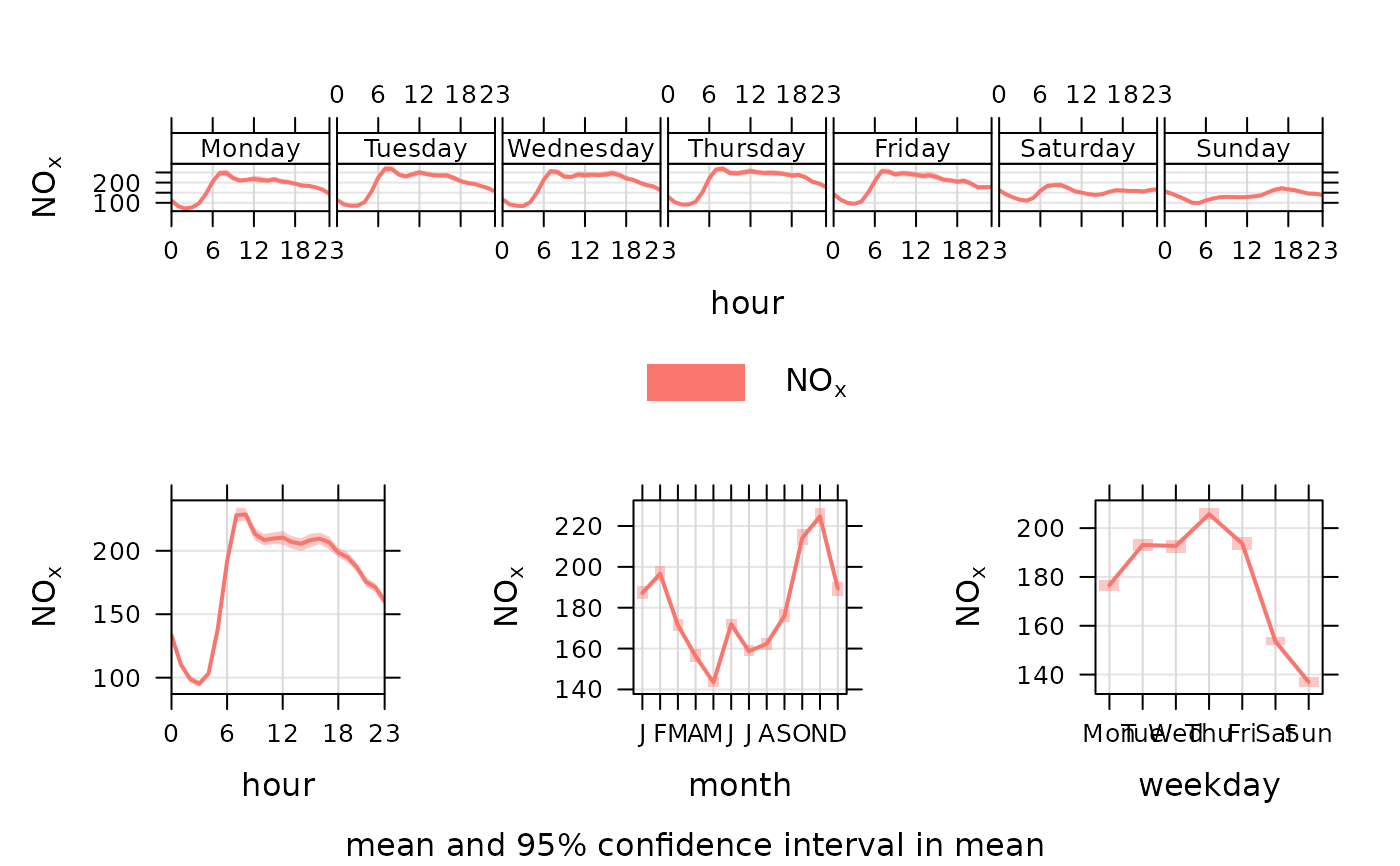 # for a subset of conditions
if (FALSE) { # \dontrun{
timeVariation(subset(mydata, ws > 3 & wd > 100 & wd < 270),
pollutant = "pm10", ylab = "pm10 (ug/m3)"
)
# multiple pollutants with concentrations normalised
timeVariation(mydata, pollutant = c("nox", "co"), normalise = TRUE)
# show BST/GMT variation (see ?cutData for more details)
# the NOx plot shows the profiles are very similar when expressed in
# local time, showing that the profile is dominated by a local source
# that varies by local time and not by GMT i.e. road vehicle emissions
timeVariation(mydata, pollutant = "nox", type = "dst", local.tz = "Europe/London")
# In this case it is better to group the results for clarity:
timeVariation(mydata, pollutant = "nox", group = "dst", local.tz = "Europe/London")
# By contrast, a variable such as wind speed shows a clear shift when
# expressed in local time. These two plots can help show whether the
# variation is dominated by man-made influences or natural processes
timeVariation(mydata, pollutant = "ws", group = "dst", local.tz = "Europe/London")
# It is also possible to plot several variables and set type. For
# example, consider the NOx and NO2 split by levels of O3:
timeVariation(mydata, pollutant = c("nox", "no2"), type = "o3", normalise = TRUE)
# difference in concentrations
timeVariation(mydata, poll = c("pm25", "pm10"), difference = TRUE)
# It is also useful to consider how concentrations vary by
# considering two different periods e.g. in intervention
# analysis. In the following plot NO2 has clearly increased but much
# less so at weekends - perhaps suggesting vehicles other than cars
# are important because flows of cars are approximately invariant by
# day of the week
mydata <- splitByDate(mydata, dates = "1/1/2003", labels = c("before Jan. 2003", "After Jan. 2003"))
timeVariation(mydata, pollutant = "no2", group = "split.by", difference = TRUE)
# sub plots can be extracted from the openair object
myplot <- timeVariation(mydata, pollutant = "no2")
plot(myplot, subset = "day.hour") # top weekday and plot
# individual plots
# plot(myplot, subset="day.hour") for the weekday and hours subplot (top)
# plot(myplot, subset="hour") for the diurnal plot
# plot(myplot, subset="day") for the weekday plot
# plot(myplot, subset="month") for the monthly plot
# numerical results (mean, lower/upper uncertainties)
# myplot$data$day.hour # the weekday and hour data set
# summary(myplot, subset = "hour") #summary of hour data set
# head(myplot, subset = "day") #head/top of day data set
# tail(myplot, subset = "month") #tail/top of month data set
# plot quantiles and median
timeVariation(mydata, stati = "median", poll = "pm10", col = "firebrick")
# with different intervals
timeVariation(mydata,
stati = "median", poll = "pm10", conf.int = c(0.75, 0.99),
col = "firebrick"
)
# with different (arbitrary) panels
# note 'hemisphere' is passed to cutData() for season
timeVariation(
mydata,
pollutant = "no2",
panels = c("weekday.season", "year", "wd"),
hemisphere = "southern"
)
} # }
# for a subset of conditions
if (FALSE) { # \dontrun{
timeVariation(subset(mydata, ws > 3 & wd > 100 & wd < 270),
pollutant = "pm10", ylab = "pm10 (ug/m3)"
)
# multiple pollutants with concentrations normalised
timeVariation(mydata, pollutant = c("nox", "co"), normalise = TRUE)
# show BST/GMT variation (see ?cutData for more details)
# the NOx plot shows the profiles are very similar when expressed in
# local time, showing that the profile is dominated by a local source
# that varies by local time and not by GMT i.e. road vehicle emissions
timeVariation(mydata, pollutant = "nox", type = "dst", local.tz = "Europe/London")
# In this case it is better to group the results for clarity:
timeVariation(mydata, pollutant = "nox", group = "dst", local.tz = "Europe/London")
# By contrast, a variable such as wind speed shows a clear shift when
# expressed in local time. These two plots can help show whether the
# variation is dominated by man-made influences or natural processes
timeVariation(mydata, pollutant = "ws", group = "dst", local.tz = "Europe/London")
# It is also possible to plot several variables and set type. For
# example, consider the NOx and NO2 split by levels of O3:
timeVariation(mydata, pollutant = c("nox", "no2"), type = "o3", normalise = TRUE)
# difference in concentrations
timeVariation(mydata, poll = c("pm25", "pm10"), difference = TRUE)
# It is also useful to consider how concentrations vary by
# considering two different periods e.g. in intervention
# analysis. In the following plot NO2 has clearly increased but much
# less so at weekends - perhaps suggesting vehicles other than cars
# are important because flows of cars are approximately invariant by
# day of the week
mydata <- splitByDate(mydata, dates = "1/1/2003", labels = c("before Jan. 2003", "After Jan. 2003"))
timeVariation(mydata, pollutant = "no2", group = "split.by", difference = TRUE)
# sub plots can be extracted from the openair object
myplot <- timeVariation(mydata, pollutant = "no2")
plot(myplot, subset = "day.hour") # top weekday and plot
# individual plots
# plot(myplot, subset="day.hour") for the weekday and hours subplot (top)
# plot(myplot, subset="hour") for the diurnal plot
# plot(myplot, subset="day") for the weekday plot
# plot(myplot, subset="month") for the monthly plot
# numerical results (mean, lower/upper uncertainties)
# myplot$data$day.hour # the weekday and hour data set
# summary(myplot, subset = "hour") #summary of hour data set
# head(myplot, subset = "day") #head/top of day data set
# tail(myplot, subset = "month") #tail/top of month data set
# plot quantiles and median
timeVariation(mydata, stati = "median", poll = "pm10", col = "firebrick")
# with different intervals
timeVariation(mydata,
stati = "median", poll = "pm10", conf.int = c(0.75, 0.99),
col = "firebrick"
)
# with different (arbitrary) panels
# note 'hemisphere' is passed to cutData() for season
timeVariation(
mydata,
pollutant = "no2",
panels = c("weekday.season", "year", "wd"),
hemisphere = "southern"
)
} # }
